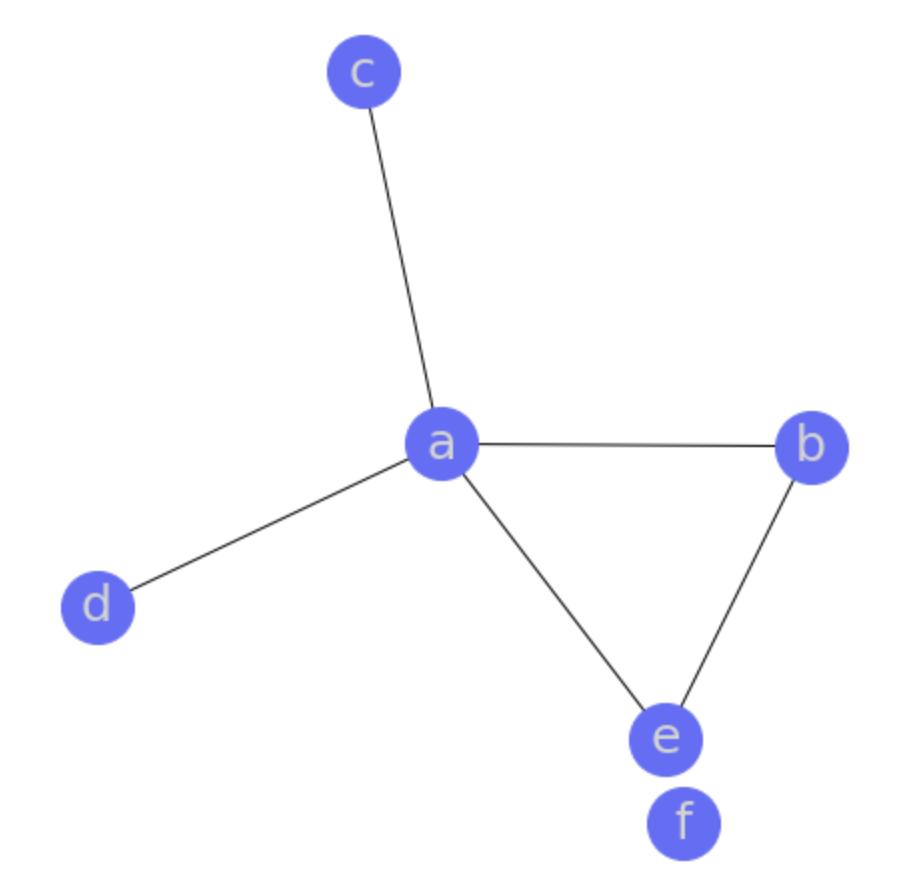
Networkx
import networkx as nx
import matplotlib.pyplot as plt
def draw_graph(graph: Any, figsize=(6,6), **kwargs):
G = nx.Graph(graph)
pos = nx.kamada_kawai_layout(G)
plt.figure(figsize=figsize)
default_kwargs = {
"node_size": 1300, "node_color": "#636EFA",
"font_size": 25, "font_color": "lightgray",
}
default_kwargs.update(kwargs)
nx.draw(G,pos, with_labels=True, **default_kwargs)
cyclic_graph = {
'a': {'c', 'd', 'e', 'b'}, 'b': {'e', 'a'}, 'c': {'a'},
'd': {'a'}, 'e': {'b', 'a'}, 'f': {},
}
draw_graph(cyclic_graph)
Plotly
Use the library (github) explained in this blogpost.
import random
import plotly.express as px
import visualize as gvz
# Optional if you want to upload to chart-studio
import chart_studio.plotly as py
def _node_color(self, *args, **kwargs):
return random.choice(px.colors.qualitative.Plotly[:3])
cyclic_graph = {
'a': {'c', 'd', 'e', 'b'}, 'b': {'e', 'a'}, 'c': {'a'},
'd': {'a'}, 'e': {'b', 'a'}, 'f': {},
}
fig = gvz.GraphVisualization(
G=nx.Graph(non_cyclic_graph),
# Describes where to place in 2-D. There are other layouts. The other one I
# recommend is nx.spring_layout(G)
pos=nx.kamada_kawai_layout(G),
node_text_position='top center',
node_color=_node_color,
node_size=25,
node_text_font_size={4: 30},
edge_color={(0, 1): '#ff0000'},
).create_figure()
fig.show()
# Only if you want to upload to chart-studio
py.plot(fig, filename='Network graph plotting in plotly', auto_open=True)
Graphviz
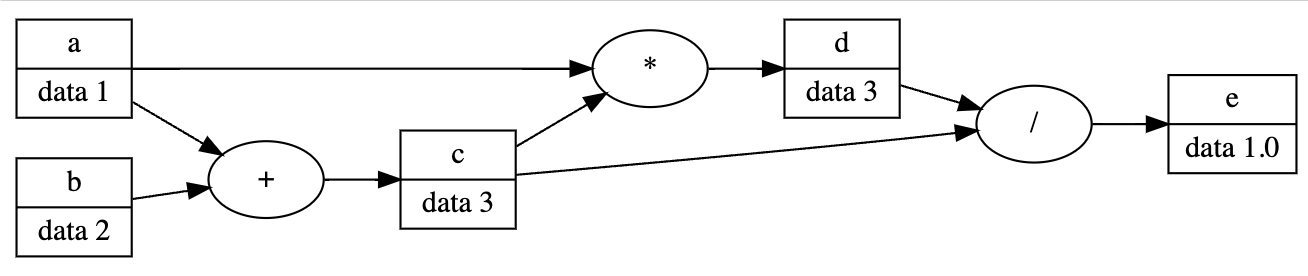
The following code is heavily inspired by Andrej Karpathy’s lectures on deep learning.
import graphviz as gv
import dataclasses as DC
import typing as T
@DC.dataclass
class Node:
name: str
val: T.Optional[T.Union[int, float, str]] = None
#: Which operation created this node. Example "+", "-", "*", "/"
#: We are setting init to false as we want to create the other nodes before
#: updating the op.
op: str = DC.field(init=False)
#: Nodes which created this node.
prev: set['Node'] = DC.field(default_factory=set)
def __hash__(self):
return id(self)
def draw_dot(root: Node, only_nodes: T.Optional[set[Node]]=None):
# LR = left to right
dot = gv.Digraph(format="svg", graph_attr={"rankdir": "LR"})
nodes, edges = _trace(root)
for n in nodes:
# for any value in the graph, create a rectangular ('record') node for it
dot.node(name=n.name, label=f"{n.name} | data {n.val}", shape="record")
if n.op:
# if this value is a result of some operation, create an op node for it
dot.node(name=n.name+n.op, label=n.op)
# and connect this node to it
dot.edge(n.name+n.op, n.name)
for n1, n2 in edges:
# connect n1 to the op node of n2
dot.edge(n1.name, n2.name+n2.op)
return dot
def _trace(root):
"""Builds a set of all nodes and edges in a graph"""
nodes, edges = set(), set()
_build_trace(v=root, nodes=nodes, edges=edges)
return nodes, edges
def _build_trace(v, nodes, edges):
if v not in nodes:
nodes.add(v)
for child in v.prev:
if child is None:
continue
edges.add((child, v))
_build_trace(v=child, nodes=nodes, edges=edges)
"""
c = a+b
d = a*c
e = c / d
"""
a = Node(name='a', val=1)
b = Node(name='b', val=2)
c = Node(name='c', val=a.val + b.val)
d = Node(name='d', val=a.val * c.val)
e = Node(name='e', val=c.val / d.val)
a.op, b.op = None, None
c.op = '+'
c.prev |= {a, b}
d.op = '*'
d.prev |= {c, a}
e.op = '/'
e.prev |= {c, d}